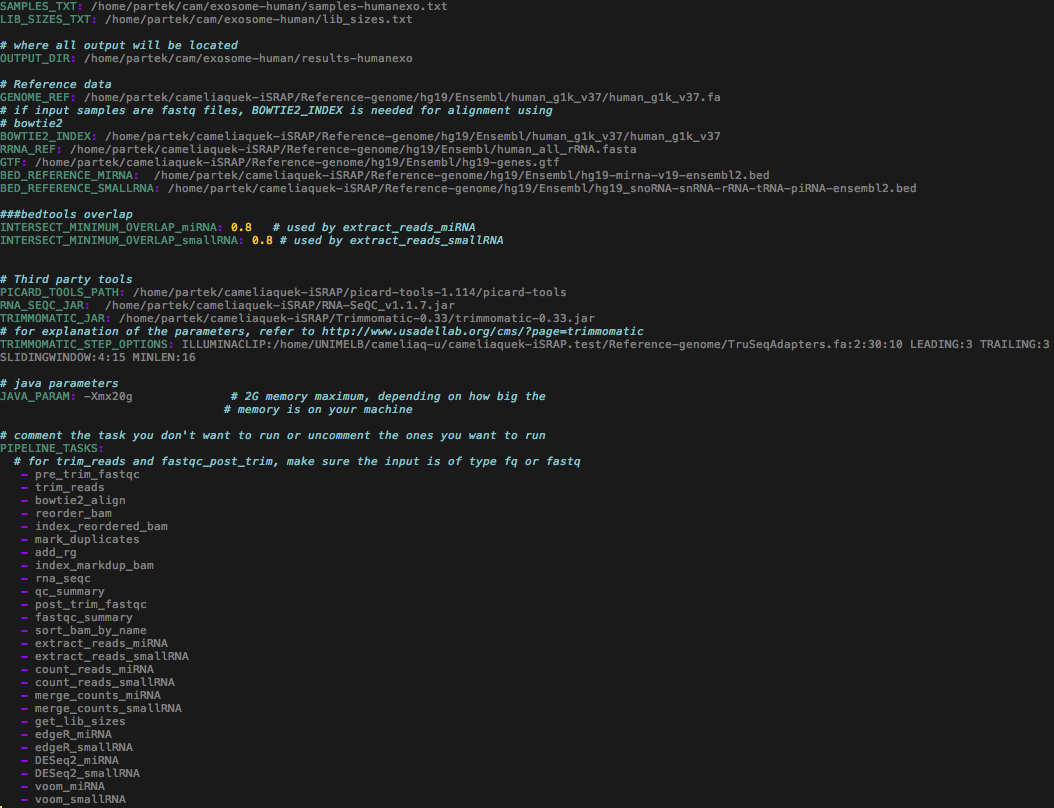
A total of 14 size-selected sequencing libraries were retrieved from Sequence Read Archive (SRA) database (Accession no. SRP034590). The miRNA samples and their replicates were extracted from plasma exosomes of seven human individuals. The focus of this previously published study (Huang et al. 2013) was to characterise exosomal RNA profiles in human plasma samples and to evaluate the reproducibility of miRNA data sets between the replicates. An example of the configuration file before running the command "python smallrna_pipeline.py":
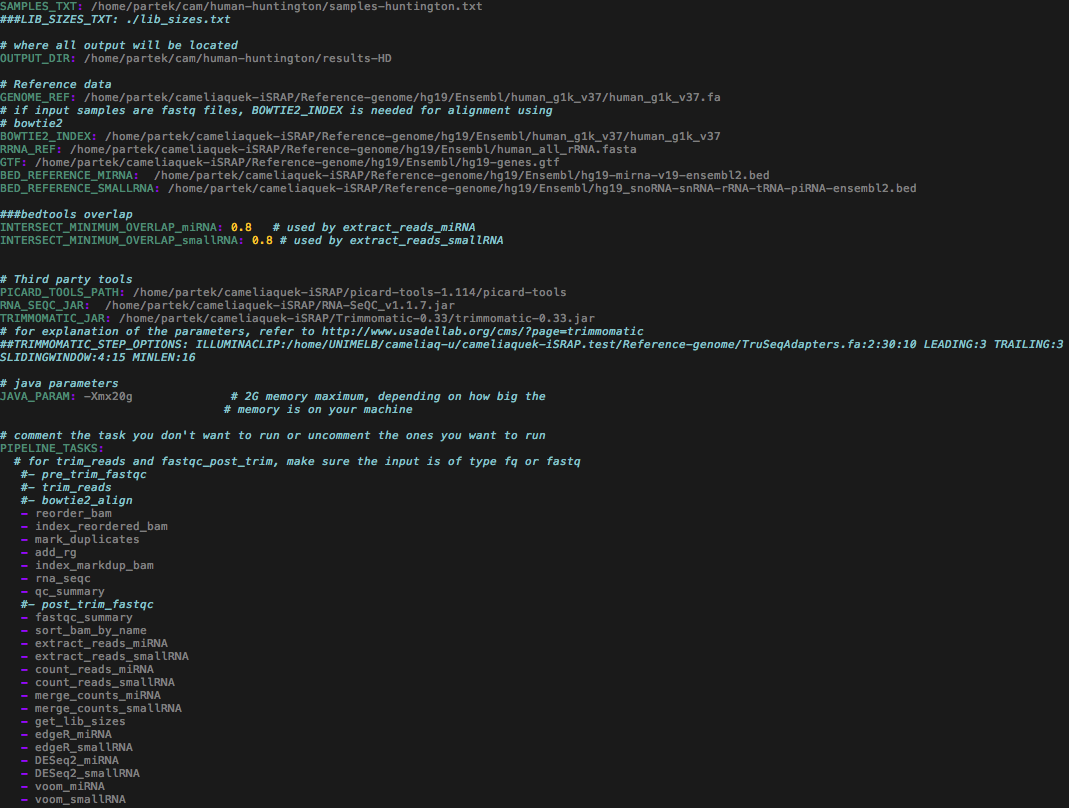
The aligned data consisted of small RNA (miRNA) sequencing libraries of HD patients (n = 12) and healthy controls (n = 11) obtained from the prefrontal cortex Brodmann Area 9 of the brain tissue (Accession no. ERP004592). Part of the scope of this published study (Hoss et al. 2014) was to investigate the presence of altered miRNA expression in HD. The setting for this study was similar to "case study 1". However, specific bioinformatic tasks were selected as the data were already aligned to the reference genome.
Under construction...